Characterization of an eye field-like state during optic vesicle organoid development

Andrew Papanastasiou

Liusaidh Owen
By Andrew Papanastasiou and Liusaidh Owen
Andrew Papanastasiou and Liusaidh Owen take us on a deep dive into their study at the MRC Human Genetics Unit.
The eye field, its development and challenges in understanding
The establishment or `specification’ of the eye field marks the first known stage of vertebrate eye development. This crucial event occurs in the developing forebrain and is defined by a group of seemingly indistinguishable cells starting to express a specific set of genes known as transcription factors (TFs). Expression of the eye-field transcription factors (EFTFs) endows these cells and their progeny with the potential to eventually form complex structures of the eye such as the retinal pigment epithelium, the neural retina and the optic stalk.
Most of our current understanding of the eye field has come from non-mammalian model systems, and in particular Xenopus Laevis, the African Clawed frog. Because this important event in development happens so early in mammals (during gastrulation, approximately embryonic day 7 in mouse, and day 18 in human embryos), and at this stage the tissue relevant for the study of the eye field is both tiny in volume as well as extremely challenging to harvest, progress in understanding this very early event in mammalian eye development has not been possible using in vivo approaches.
Clinical genetic studies (including past seminal studies by researchers at the MRC Human Genetics Unit) have shown that mutations in genes such as PAX6, RAX, OTX2 and SIX3 – members of the set of EFTFs identified in Xenopus – can lead to severe developmental eye malformations. Identifying whether the same genes identified in non-mammalian studies, are involved in mammalian eye-field specification, and furthermore, what the role these genes play in stably establishing the eye field, is both fundamental for understanding human eye development as well as an important step to better informing genetic diagnoses in the clinic.
The study
Our overarching goal in this study was to exploit an ex vivo model of mammalian early eye development (mouse stem-cell derived optic vesicle organoids) to gain an in depth understanding of the key transitions that cells must go through to acquire the eye-field identity.
We had three specific aims. Firstly, we wanted to check whether the genes defining the eye-field in mammalian systems overlapped with those discovered in Xenopus; secondly, we wanted to gain a broader quantitative understanding of what gene-expression dynamics occur as cells acquire the optic fate; and thirdly, we wanted to gain a deeper molecular understanding of what regulates these crucial gene expression changes.
For the latter aim, we wanted both to understand which transcription factors play roles in regulation as well as what specific genomic regulatory elements (gene promoters and distal enhancers) these transcription factors act through.
Although the questions we were trying to tackle were specifically in the context of eye development, the broader question of what changes in gene expression happen during cell-fate transitions and identifying the regulatory elements and transcription factors controlling these transitions, is one that is of interest across developmental biology. This understanding may also help researchers pinpoint the causes of malformations that occur when mutations in our genome disrupt the normal cascade of events leading to eye-field specification.
We hope that the questions we began to tackle in this study will provide the basis of further insights into genetic developmental eye diseases in humans and inform clinical diagnoses.
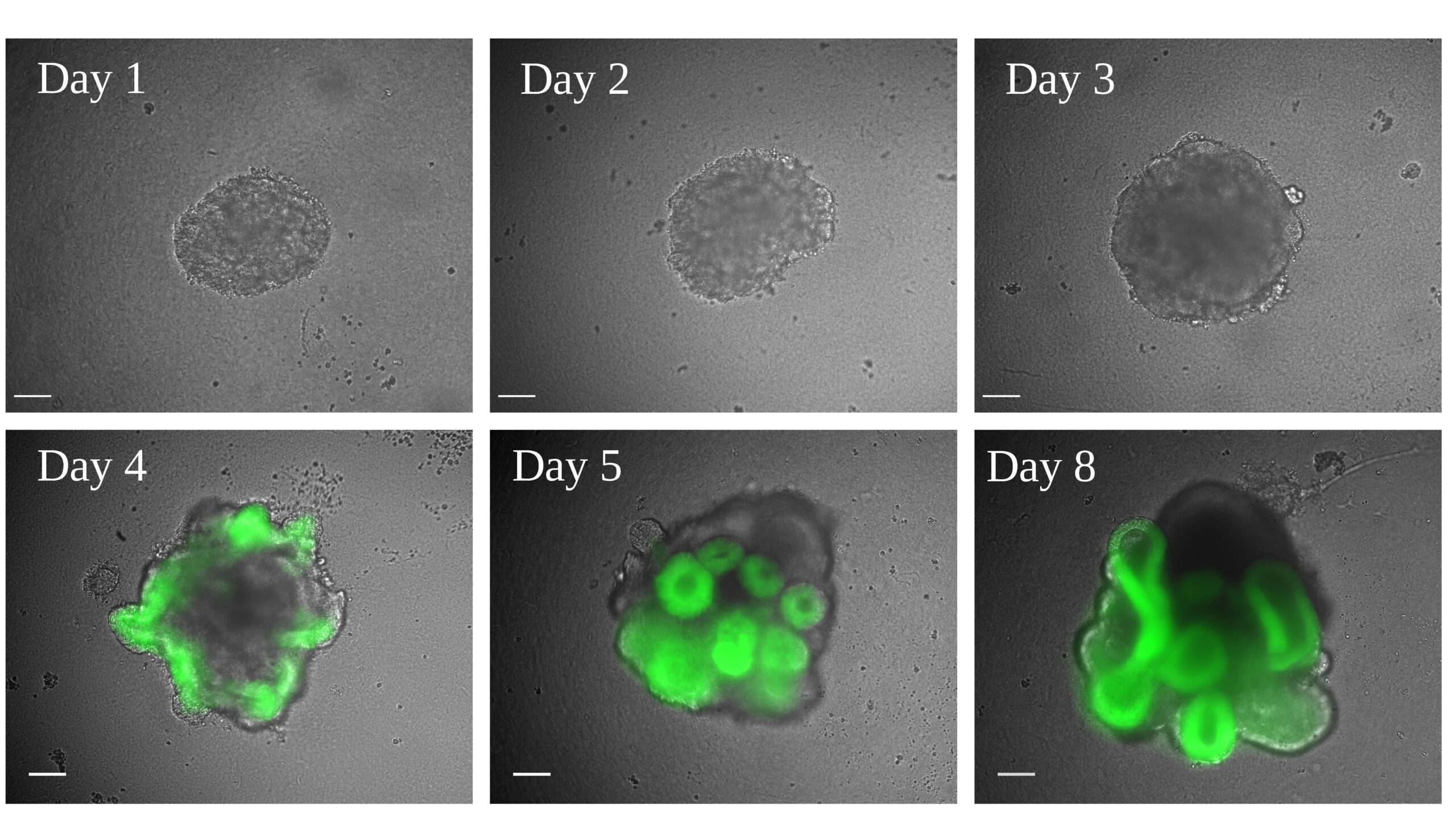
We characterised and optimised the growth, differentiation and reproducibility of an optic vesicle organoid model system, that was first described by the Sasai lab. We used a mouse embryonic stem cell line that has a GFP fluorescent reporter cassette knocked into the RAX locus. This gave us a clear read out for eye field formation.
We grew and collected cells from organoids on day 1 to day 5 of differentiation and collected RNA for bulk RNA-seq. We also used the same protocol to collect organoids for ATAC-seq that could be analysed in combination with the transcriptome data to show how changes in RNA levels correlate with chromatin accessibility.
The time course of samples meant that we could track changes in gene expression over time and identify genes that are expressed when the eye field is formed. Combining our transcription and accessibility data with thorough bioinformatic analysis we aimed to predict which DNA elements control the gene expression changes.
See above, Fig 1A. Exploiting optic vesicle organoids to probe the molecular origins of eye-field specification. (A) Representative organoids from days 1-5, and day 8. Scale bars: 100 µm
Computational analysis
We were particularly interested in understanding the dynamic patterns of gene expression occurring during the “switch-on” of the eye-field. By performing computational analyses of the transcriptomic changes across optic vesicle differentiation we found that the establishment of the eye field in mouse involves the strong up-regulation of a set of genes that includes the same key transcription factors identified in Xenopus.
Using a variety of bioinformatic analyses together with statistical modelling approaches, we developed a framework to systematically integrate different data modalities (transcriptomics and chromatin-accessibility) that helped us gain valuable insights into the regulation of these gene-expression dynamics.
We showed that the key EFTFs, and in particular RAX, PAX6 and LHX2, likely play a crucial role in regulating the stable transition of cells to the ocular fate, by binding cis-regulatory elements (`enhancers’ and `repressors’) in genomic domains local to EF genes. By computationally linking changes in accessibility at individual genomic loci to these key TFs, we generated hypotheses of plausible cis-regulatory elements required for the differentiation from pluripotency to eye field cell states.
Experimental validation
We wanted to experimentally validate our strategy for identifying cis-regulatory elements by testing the influence of our predicted elements on optic vesicle differentiation. We selected two of the EFTFs, RAX and SIX6, and chose one representative predicted cis-regulatory region for each gene that had chromatin accessibility and TF binding changes most indicative of these regions being involved in optic vesicle differentiation.
Using CRISPR we introduced deletions encompassing these regions and then differentiated the resulting cell lines. RNA-seq on these mutant organoids revealed a modest but significant decrease in the expression of the target gene and other EFTFs. This supports our approach to defining cis-regulatory elements and demonstrates the regulatory potential of two new DNA regions in controlling gene expression changes important to optic vesicle formation.
This approach does not have the resolution to study the mechanisms that trigger the initial expression of the EFTFs. We therefore broadened our approach to address this unknown by integrating publicly available datasets and using these to formulate a model by which key developmental genes may be switched on. We propose that a change in the SOX2 binding partner from POU5f1 to OTX2 at one of our predicted cis-regulatory regions is the initial trigger for RAX expression and have laid out the experiments necessary to validate.
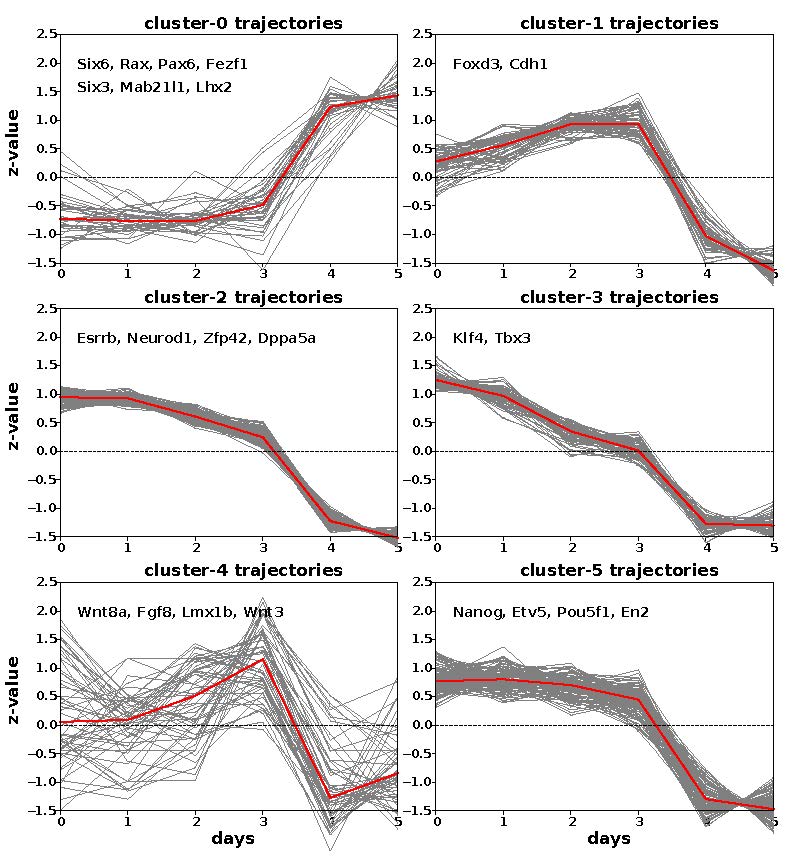
Fig 2B. Clustering of EF-up and EF-down gene trajectories reveals six clusters with three underlying patterns across the time course. At each time point, heatmap and trajectory plots indicate expression averaged across triplicates.
Conclusion
To summarise, we have characterised the gene expression and chromatin accessibility changes associated with eye field specification in mammals and collated a new list of mammalian EFTFs. We also developed a framework for predicting regions of DNA with regulatory activity and experimentally demonstrated the effect of removing these on gene expression.
Significance of the study
The work we performed has extended the early characterization of the eye-field to mammalian model systems and furthermore, the unique data that were generated together with the computational analysis framework we put together to analyse these, have provided key insights on the regulatory roles of key EFTFs during this crucial stage of eye development.
Eye-field specification in humans is currently impossible to study in vivo or from donated tissues. As such, we envision that performing similar experiments and analyses using human derived OV-organoids, can help identify key molecular mechanisms operating during earliest stage of human eye development and therefore push this work closer to having translational potential.
Read the paper, Characterization of an eye field-like state during optic vesicle organoid development published in Development.



