Single cell epigenome landscape of development and ageing

In January, Wolf Reik came to the IGMM to speak at the weekly seminar series. Wolf is a leading scientist in the field of epigenetics. Epigenetics is the study of changes in cells which affect gene activity rather than altering the actual genetic sequence. This usually involves some kind of reversible chemical modification to the DNA. These changes are particularly relevant in animal development, which is what Wolf’s lab primarily works on.

Wolf Reik at the Lister Institute Centenary dinner in London, 1991. L-R Wendy Bickmore (now Director of the MRC HGU), Robin Allshire, Cristina Rada, Wolf Reik, Ian Jackson, Sally Cross. Picture provided by Ian Jackson.
After an entertaining introduction by Tamir Chandra, who used to work in Wolf’s lab as a postdoc, featuring an old photo of Wolf with some scientists that now work at the IGMM, Wolf began to talk about his work on gastrulation. As the biologist and author Lewis Wolpert once put it, ‘It’s not birth, marriage or death, but gastrulation that is truly the most important time in your life.’ Gastrulation is a process during animal development where each of the cells within the early embryo become programmed to develop into different organs. During this process, 40 identical-looking cells expand to become 40,000 more specialised ones, and the embryo begins to look significantly more like an animal. Although this process is clearly very important in development, we do not yet understand how it works at a molecular level.
Together with a diverse group of collaborators spanning the fields of statistics, machine learning and developmental biology, Wolf’s lab generated an ‘atlas’ of mouse gastrulation using single cell transcriptomic data from 116,312 cells. This was done before the advent of high throughput droplet single cell sequencing systems, so is an impressive achievement, and the resultant atlas can be freely accessed online. However, this atlas did not give information on the molecular mechanisms underlying how the three characteristic germ layers are established, which takes place during gastrulation. These are the ectoderm, which gives rise to the skin and the nervous system, the mesoderm, which develops into bone and muscle, and the endoderm, which forms organs such as the liver and pancreas, and also makes up the linings of the digestive and respiratory system.
Wolf’s lab therefore set out to understand the epigenetic changes underlying germ layer specification, using a technique they developed called scNMT-seq (single-cell Nucleosome, Methylome and Transcriptome sequencing). This allows data on chromatin accessibility, DNA methylation, and transcription to all be collected from one single cell. This is particularly useful in the study of enhancers. Enhancers are short regions of DNA involved in gene regulation, which usually increase the activity of their target genes. Development of each of the different germ layers has its own associated enhancers, which become demethylated in the cells that eventually differentiate into each corresponding layer. Using scNMT-seq, Wolf’s lab found that the ectodermal enhancers become demethylated at an earlier stage than the mesodermal and endodermal enhancers. This could explain why in the lab, the default differentiation state of stem cells is to become neural tissue, which comes from the ectoderm layer. Now they are working on improving the time resolution of this technique, both computationally and in the lab, so they can find out at what exact point in development this demethylation takes place. In addition, Wolf is part of the new Wellcome-funded Human Developmental Developmental Biology Initiative, which will use the same single-cell methods to analyse cells from human embryos, investigating whether the same regulatory mechanisms apply in humans.
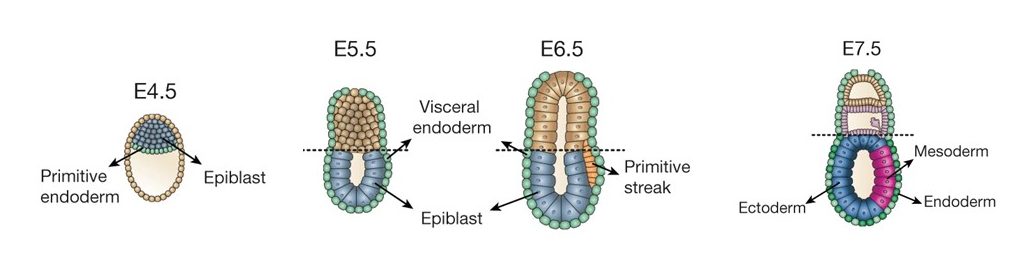
The stages of mouse development used in the scNMT-seq study. Gastrulation takes place between E6.5 and E8.5. Image adapted from Argelaguet et al, 2019.
Next Wolf went on to talk about the lab’s work on DPPA2 and 4, two genes which are expressed in very early stage embryos but not in any adult tissues. However, mice without either gene survive past this early stage, before dying around birth from organ defects. As well as enhancers, which I mentioned earlier, gene expression can also be regulated by factors binding just upstream of the start of the gene, in a region known as the promoter. In the very early stage embryos, the promoters for many genes are bound by both activating and repressing factors in a state known as bivalency. Wolf’s lab found that DPPA2 and 4 bind to these bivalent promoters and also that without DPPA2 and 4, bivalency is lost. This explains why the mice without these genes do not show a phenotype until birth although DPPA2 and 4 are not expressed past the very early stage – genes with these bivalent promoters are unable to be activated due to the loss of bivalency, and these genes may be required much later in development.
The talk ended with Wolf tackling one of life’s exciting questions – can we reverse ageing? DNA methylation changes with age and as a result we can tell someone’s age (within 3.6 years) by looking at methylation of DNA in their blood cells. You may have heard of induced pluripotent stem cells (iPS cells), a type of stem cell which can be generated from cells in our body using specific factors. The discovery of iPS cells won Shinya Yamanaka the Nobel Prize in 2012. Taking fibroblasts and converting them into iPS cells causes the methylation clock to go back to zero. Wolf’s lab have been able to partially reprogram these same cells so their methylation reflects a specific age. Perhaps one day we will be able to do this on cells in our own bodies, enabling us to revisit a specific age – a distant but intriguing prospect.
References & Further Reading
Argelaguet R, Clark SJ, Mohammed H, Stapel LC, Krueger C, Kapourani CA, Imaz-Rosshandler I, Lohoff T, Xiang Y, Hanna CW, Smallwood S, Ibarra-Soria X, Buettner F, Sanguinetti G, Xie W, Krueger F, Göttgens B, Rugg-Gunn PJ, Kelsey G, Dean W, et al (2019) Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 576: 487–491
Clark SJ, Argelaguet R, Kapourani CA, Stubbs TM, Lee HJ, Alda-Catalinas C, Krueger F, Sanguinetti G, Kelsey G, Marioni JC, Stegle O & Reik W (2018) scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9: 1–9
Eckersley-Maslin MA, Parry AJ, Blotenburg M, Krueger C, Franklin VNR, Clark S, D’Santos CS & Reik W (2019) Dppa2/4 target chromatin bivalency enabling multi-lineage commitment. bioRxiv: 832873
Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, Mulas C, Ibarra-Soria X, Tyser RCV, Ho DLL, Reik W, Srinivas S, Simons BD, Nichols J, Marioni JC & Göttgens B (2019) A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566: 490–495



