Nanopore Sequencing: Controversial World Records and Whale Watching
Back in January, Prof. Matt Loose of the University of Nottingham gave a seminar at the IGMM. As a sequencing tech junkie, I was very excited to hear him speak as Matt works closely with Oxford Nanopore Technologies (ONT) delving into the potential applications of their long read sequencing platforms. Having previously worked at Illumina I was well aware of ONT, but Matt’s seminar revealed to me just how powerful their technology can be.
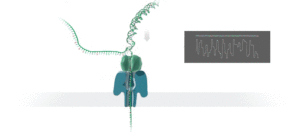
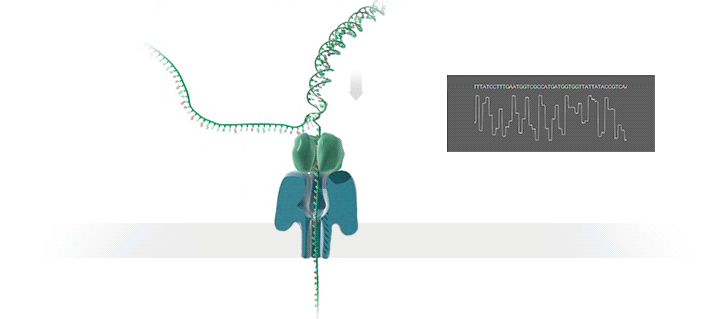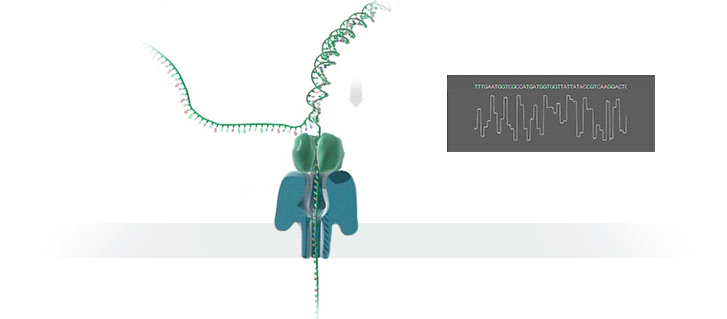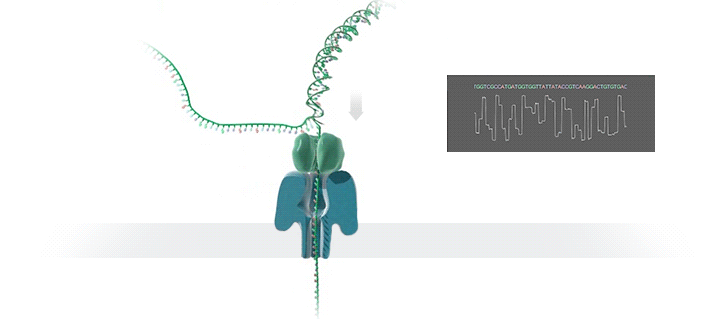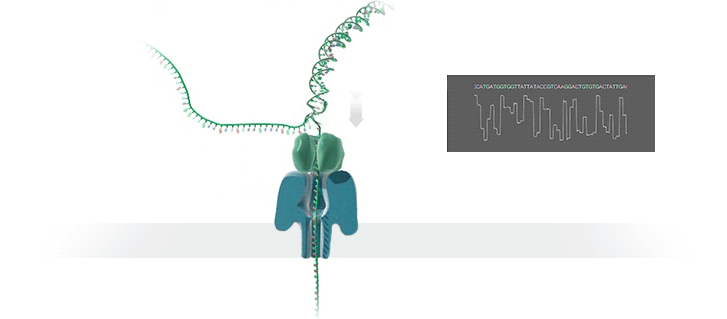
Figure 1 Nanopore sequencing Source: https://nanoporetech.com/how-it-works
ONT platforms utilise nanopore sequencing, where a protein nanopore is embedded in a membrane and a voltage difference applied across it. DNA travels through the nanopore causing changes in this voltage as it does. The changes are detected and read by software which converts the signal into the As Ts Gs and Cs that make up DNA.
Matt has been exploring nanopore sequencing for the past few years, and is so dedicated to the cause he and fellow collaborators have set up a Long Read Club; a hub for those devoted to exploring boundaries of nanopore sequencing. They are particularly active on Twitter so it’s worthwhile heading there for the latest updates if you’re interested.

The Long Read Club’s whale mascot source: https://www.longreadclub.org/
The ‘Whale Scale’ is something the Long Read Club has devised to try to illustrate just how massive the reads they are generating are; converting read length in kilobases to a comparatively large whale as a representative. This ‘whale watching’ introduces a competitive streak to the Long Read community, with an ashes trophy being passed between labs as they race to outdo each other in sequencing the longest single read. Currently this trophy resides in Matt’s office as his lab successfully produced a single read of just over 2.3 megabases in length. The record is controversial though, as the read is technically 11 separate reads, which Matt defends as a software glitch. Regardless, the team at Circulomics recently published a 2.4 megabase read, meaning Matt will have to relinquish his hold on the trophy, controversies aside.
Matt spoke a lot through his seminar on the development work he and others have been doing to achieve these colossal read lengths. With ONT platforms read length is theoretically infinite, the limiting factor being the length of the input DNA molecule itself. But preserving native DNA length is no easily achievable feat. Source, library preparation and careful sample handling all come into play in determining read length. The dedication Matt’s team had in achieving their record read length was illustrated when he recounted how they pipetted samples painstakingly slowly – taking 10 minutes to complete a single pipetting motion; a feat not everyone could be so diligent and fearless as to undertake. Matt also emphasised the importance of updated basecalling software in improving sequence quality and error rates, highlighting the many areas of expertise needed to make this technology work.
But with all this effort to make reads longer, what extra information can we actually gleam from longer reads that we can’t already obtain from pre-existing technologies?
Matt highlighted the many weaknesses of traditional short read technologies which nanopore sequencing combats, offering direct sequencing of RNA (Workman et al., 2018) as well as detection of DNA methylation patterns (Rand et al., 2017; Simpson et al., 2017). Long read data from ONT devices has helped fill gaps in many reference genomes. Matt himself was involved in the project that generated the most comprehensive human reference genome to date (Jain et al., 2018) where ONT sequencing was used to patch with pre-existing short read data to fill gaps that had previously gone unfilled – mostly highly repetitive regions of the genome including the MHC locus. Long reads can also be used to estimate the lengths of telomeres, and recently the first ever telomere to telomere assembly of a human chromosome was announced (Miga et al., 2019).

Figure 3. ONT’s handheld minION device. source:
For me though, it’s the portability of ONT’s platforms that stands out the most. Anyone who has stepped inside a sequencing lab to see a mammoth HiSeq plonked on top of a bench can agree that conventional sequencing methods are limited in their accessibility. In contrast,ONT’s handheld minION device (pictured) has the honour of being the first sequencing platform to be used in space (Castro-Wallace et al., 2017).
And in the face of the current Covid-19 pandemic it is not difficult to understand the invaluable benefits of rapid and accurate onsite sequencing, saving immeasurable amounts of both time and money in the face of tracing the spread of a novel disease.
ONT have their sights on making sequencing even more portable as they’re currently developing the SmidgION; a sequencing device that could attach to your mobile phone, widening the accessibility of DNA sequencing and analysis to more people and more locations.
The contributions ONT’s platforms are making to the sequencing world are certainly invaluable however, their low read accuracy remains a major issue with the technology. Error rates range from 5-15% (Rang, Kloosterman, & de Ridder, 2018), and reads often need to be confirmed via complimentary methods. With people like Matt and the other members of the Long Read Club working on it however, I don’t think it will be too long before this obstacle is surmounted.
Castro-Wallace, S. L., Chiu, C. Y., John, K. K., Stahl, S. E., Rubins, K. H., McIntyre, A. B. R., … Burton, A. S. (2017). Nanopore DNA Sequencing and Genome Assembly on the International Space Station. Scientific Reports. https://doi.org/10.1038/s41598-017-18364-0
Jain, M., Koren, S., Miga, K. H., Quick, J., Rand, A. C., Sasani, T. A., … Loose, M. (2018). Nanopore sequencing and assembly of a human genome with ultra-long reads. Nature Biotechnology. https://doi.org/10.1038/nbt.4060
Miga, K. H., Koren, S., Rhie, A., Vollger, M. R., Gershman, A., Bzikadze, A., … Phillippy, A. M. (2019). Telomere-to-telomere assembly of a complete human X chromosome. BioRxiv. https://doi.org/10.1101/735928
Rand, A. C., Jain, M., Eizenga, J. M., Musselman-Brown, A., Olsen, H. E., Akeson, M., & Paten, B. (2017). Mapping DNA methylation with high-throughput nanopore sequencing. Nature Methods. https://doi.org/10.1038/nmeth.4189
Rang, F. J., Kloosterman, W. P., & de Ridder, J. (2018). From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biology. https://doi.org/10.1186/s13059-018-1462-9
Simpson, J. T., Workman, R. E., Zuzarte, P. C., David, M., Dursi, L. J., & Timp, W. (2017). Detecting DNA cytosine methylation using nanopore sequencing. Nature Methods. https://doi.org/10.1038/nmeth.4184
Workman, R., Tang, A., Tang, P., Jain, M., Tyson, J., Zuzarte, P., … Timp, W. (2018). Nanopore native RNA sequencing of a human poly(A) transcriptome. BioRxiv. https://doi.org/10.1101/459529



