An Important Chap – The ChAHP Complex
On the 6th of November 2019, the IGMM welcomed Professor Marc Bühler from the Friedrich Miescher Institute for Biomedical Research to present his work on the ChAHP complex. Research in the Bühler lab focuses on investigating various mechanisms that affect eukaryotic genome organization, chromatin structure, and gene expression, and how these mechanisms have evolved over time.
The contents of Prof Bühler’s talk aimed to answer three main questions regarding the ChAHP complex:
- what does it do?
- how does the complex do it?
- what are its links to disease?
In an investigation into ADNP function, it was established that the proteins ADNP, HP1 and CHD4 formed a stable complex, and the lab also found the ChAHP complex to be important in the specification of cell fates and differentiation. The first step taken to elucidate the unknown role of ChAHP was to generate Adnp knock-out mouse embryonic stem cells. Adnp knock-out cells unexpectedly differentiated towards an endodermal lineage, with an upregulation in genes often activated upon exit from pluripotency. So, it was likely that this complex was crucial in restricting the expression of lineage-specifying genes, ensuring appropriate cell fate decisions later on.
Whilst the ChAHP complex was now known to have a role in regulating gene expression, the mechanisms of this regulation were still unknown. Perhaps the HP1 protein within the complex directed ChAHP towards sites of H3K9 methylation, leading to gene repression in heterochromatin? This original prediction was unlikely to be the case as a ChIP-seq analysis indicated that ADNP binding sites did not correlate with heterochromatin marks and were actually enriched in euchromatin instead. It was subsequently discovered that targeting of the ChAHP complex to euchromatin was mediated by ADNP, in a sequence-specific manner.
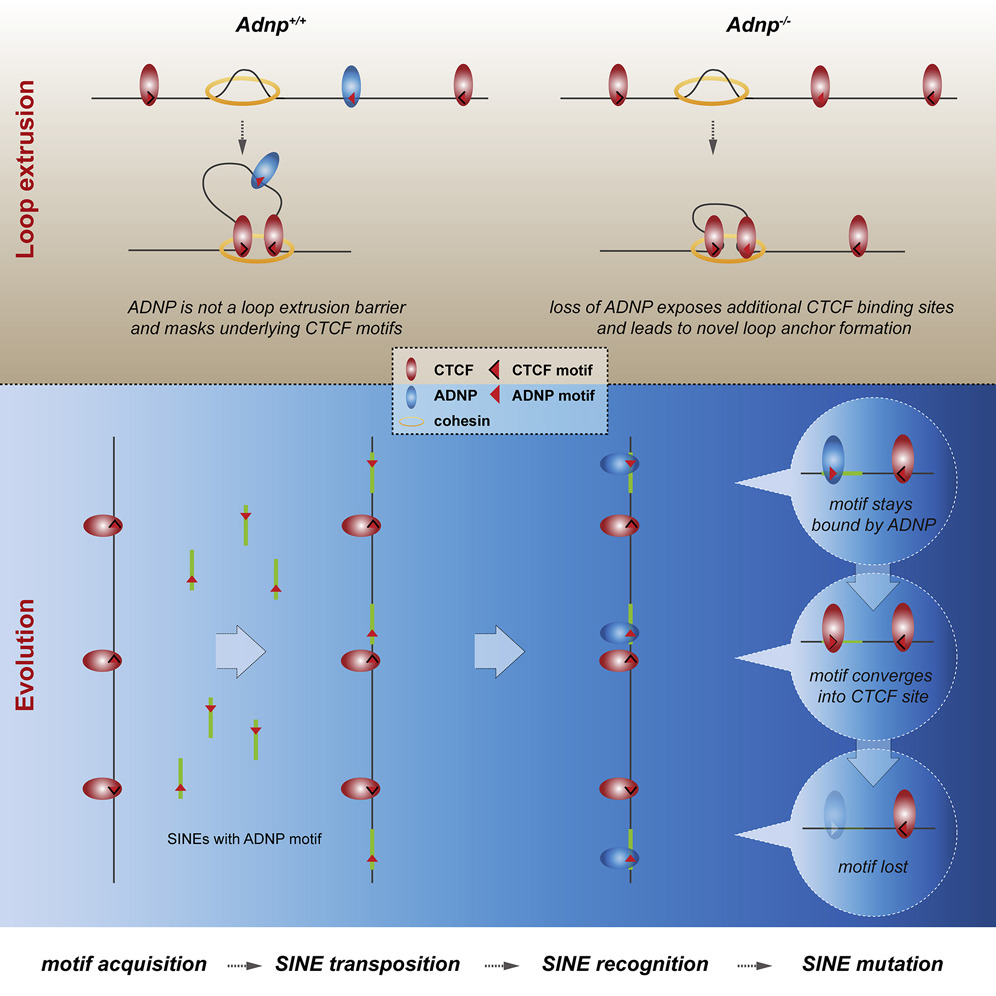
Interestingly, even though the ChAHP complex was found to repress gene activity if bound at a promoter, a majority of ADNP peaks from the ChIP-seq analysis were not localized proximal to promoters. When revisiting these ChAHP binding sites, the Bühler lab discovered that the consensus binding site of ChAHP was surprisingly similar to that of CTCF – an important insulator, which functions by defining the boundaries of chromatin loops, thus playing a key role in regulating gene expression. By investigating CTCF peaks at ADNP binding sites in Adnp knock-out cells, ADNP was found to counteract CTCF binding at more than 15,000 sites in a competitive manner. Additionally, whilst Hi-C experiments revealed that the loss of ADNP did not affect global chromatin interactions, it was noted that approximately 100 topologically-associated domains (TADs) around ADNP binding sites had split into two smaller TADs. These results led to the conclusion that binding of the ChAHP complex prevented CTCF binding, thus inhibiting chromatin loop formation at these DNA binding sites. The importance of ChAHP in this context is clear – alterations in chromatin looping could potentially affect interactions between promoters and enhancers, thus influencing gene expression, and ultimately modify differentiation and developmental processes.
If the differential binding of ADNP and CTCF is so important, and both proteins have a similar DNA sequence preference, your next question will probably be – “how do these binding sites differ between both proteins then?” Whilst the exact answer to this question is yet to be elucidated, the SINE B2 transposable element, which has previously been linked to CTCF binding sites, may provide us with a clue. More than 90% of ADNP-bound sites overlapped with a SINE B2 element, compared to only 38% of CTCF binding sites. Furthermore, when investigating these SINE B2 sequences, a distinct preference was revealed, in which SINEs that underlay ADNP binding sites were comprised of ‘evolutionarily young’ sequences where SINE B2 sequence divergence was minimal. Prof Bühler made a fascinating speculation that the ChAHP complex may have evolved as a genomic defense mechanism, ensuring that the spread of the CTCF binding motif would be counteracted by ChAHP complex binding, preventing the immediate rewiring of genome organization by CTCF binding, which could be detrimental to organismal development.
Overall, the work by the Bühler lab has revealed the possible mechanisms and importance regarding the novel chromatin regulatory ChAHP complex. It is crucial to note that mutations in the Adnp gene have been linked to Helsmoortel-Van der Aa syndrome – a neurological disorder associated with autism spectrum disorder, caused by loss of the interaction between a truncated ADNP protein and HP1. It will be exciting to see whether the outcomes of the research carried out by the Bühler lab could translate from mice to humans, and if human endogenous retroviruses confer a similar binding of ChAHP as SINE B2 elements in mice. Hopefully the work revolving around the ChAHP complex will not only be of interest to those within the genome regulation field, but also have a clinical significance in understanding the relationship between Adnp and Helsmoortel-Van der Aa syndrome.
(Kaaij et al., 2019)
References:
This blog post was written based on the work of the Bühler lab, with specific focus on the following publications –
- Ostapcuk, V. et al. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature 557, 739–743 (2018).
- Kaaij, L. J. T., Mohn, F., van der Weide, R. H., de Wit, E. & Bühler, M. The ChAHP Complex Counteracts Chromatin Looping at CTCF Sites that Emerged from SINE Expansions in Mouse. Cell 178, 1437-1451.e14 (2019).



