Welcome!
The unano consortium exists to advance bionanotechnology research in Europe, having been kick-started by funding from the Una Europa alliance. Unano runs a monthly webinar series about bionanostructures and bionanomachines, enabling researchers across the globe to give talks on their current endeavours in the field. Anyone can attend our webinars, and the joining link and webinar programme are provided below.
The unano webinar series is co-hosted by Dr Katherine Dunn’s research group at the University of Edinburgh and Prof Jonathan Heddle’s group at Durham University. Lucy Epton at Durham is in charge of devising the webinar schedule, and if you are interested in giving a talk for us please contact her by email at lucy.epton@durham.ac.uk. James Dodd at Edinburgh is in charge of maintaining the blog and running the webinar platform and can be reached by email at J.Dodd-3@sms.ed.ac.uk.
January 2025 marked the kick-off of the unano webinar series, which expands on the previous webinar series run by Katherine’s group on the narrower theme of DNA Nanotechnology. That series was chaired and organised by Dr Matthew Aquilina initially and later James Dodd; more details (including recordings of some sessions) can be found here.
Next webinar: 10am (UK time), 25th February 2026

Join Here
Prior registration is not required. To join the webinar just click the link as (or shortly before) the webinar starts and type the name by which you wish to be known. If you join the webinar as a group please indicate this in your name, so we have some idea how big the audience is.
Webinar schedule
3pm, 29th January 2025:
James Dodd (University of Edinburgh): Title TBC
Greg Knight (Durham University): Title TBC
3pm, 26th February 2025:
Ho Yeung Chim (Ludwig-Maximilians-Universität München): Computational design of small molecule-dependent cyclic oligomers:
Designing protein oligomers that respond to small molecules is a significant challenge in dynamic protein assembly design and is crucial for interfacing with biological systems. I have introduced a new module within RFdiffusion for specifically designing cyclic oligomers by leveraging pre-existing interfaces. This design module focuses on the scaffolding of symmetrized target interfaces, ensuring that the properties of the interface are preserved in the final oligomer design. I computationally designed cyclic oligomers with varying symmetries derived from neomorphic PPIs, as a first step towards creating controllable and dynamic protein assemblies.
Göktuğ Aba (Leiden University Medical Centre): Inducing cancer cell killing using DNA nanostructure-mediated superclustering of death-receptors:
Clustering of type-II tumour necrosis factor receptors (TNFRs) is required to induce intracellular signaling. Current methods for receptor clustering lack precise control over ligand valency and spatial organization, potentially limiting optimal TNFR activation, biological insight, and therapeutic efficacy. DNA nanostructures provide nanometer-precise control over molecular arrangement, allowing control of both ligand spacing and valency. Here, we produce a DNA nanostructure decorated with controlled numbers of engineered single-chain TNF-related apoptosis-inducing ligand (sc-TRAIL) trimers. These trimers cluster death receptor 5 (DR5), enabling investigation of the geometric parameters influencing apoptotic pathway activation. We show that cell killing is affected by both valency and separation of sc-TRAIL trimers, which can be utilized to induce cell killing in human primary pancreatic and colorectal cancer organoids. Together, our data shows that precise control of receptor clustering through spacing and valency enhances our understanding of receptor activation mechanisms and informs the development of more effective cancer therapies.
3pm, 26th March 2025:
Mai P. Tran (Max Planck Institute):
Bottom-up synthetic biology seeks to engineer a cell from molecular building blocks. With DNA nanotechnology, several building blocks, such as cytoskeletons, have been reverse-engineered. However, because they currently rely on chemical synthesis and thermal annealing, synthetic cells cannot produce these DNA nanostructures from their constituents, like nucleotides. Here, we introduce RNA origami cytoskeleton mimics as an alternative set of nucleic acid-based molecular hardware for synthetic cells, which we express directly inside of GUVs containing a DNA template and a polymerse, chemically fuelled by feeding nucleotides from the outside. We designed RNA origami tiles which fold upon transcription and self-assemble into micrometer-long 3D RNA origami nanotubes at constant temperature. We find that sequence mutations on the DNA template lead to different phenotypes of RNA origami nanotubes, including the formation of closed rings. Molecular dynamics simulations and experiments show that these phenotypic transitions can be explained by alterations in the stability of RNA secondary structures and modifications to available assembly pathways. Additionally, we achieved RNA origami cortex formation and membrane deformation caused by RNA nanotube polymerization. Altogether, this work pioneers the expression of RNA origami-based hardware towards active, evolvable, and RNA-based synthetic cells.
Zuza Pakosz-Stepien (Durham University): Tailoring pH sensitive protein cages:
The rational design and creation of novel engineered protein cages are an important area of research in synthetic biology. Such cages can serve as versatile and adaptable platforms with significant applications in biomedicine. Precisely controlling the interactions between protein subunits is crucial, as it enables the controlled assembly and disassembly critical for applications such as drug delivery. Here, we present a new approach to assembling protein cages from identical protein building blocks. In previous work we were able to form protein cages with ring-shaped TRAP proteins linked via linearly coordinated gold(I) (1). Our new method utilizes TRAP with strategically positioned metal binding sites, each capable of coordinating metal ions with higher coordination numbers. These engineered proteins can self-assemble into highly stable cages in the presence of cobalt or zinc ions. Furthermore, the cages can be disassembled on demand through external triggers like chelating agents and pH changes. Interestingly, for certain triggers, the disassembly is reversible, allowing the cages to reassemble when triggering conditions cease. This work offers a promising platform for developing advanced drug delivery systems and other biomedical applications.
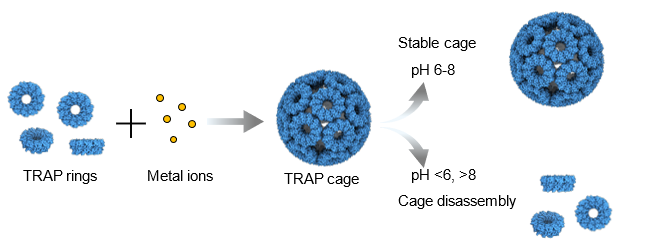
- Malay, A.D., Miyazaki, N., Biela, A. et al. An ultra-stable gold-coordinated protein cage displaying reversible assembly. Nature 569, 438–442 (2019). https://doi.org/10.1038/s41586-019-1185-4
3pm, 30th April 2025:
Sara Garcia Linares (Complutense University of Madrid): Design of nanoreactors for microplastic depolymerization based on pore-forming toxins
Actinoporins, typically non-catalytic proteins, have been engineered into biocatalytic nanoreactors capable of breaking down PET plastics. The presentation covers their mechanism of action, efficiency compared to existing PETases, and future directions such as optimizing pore size, integrating multiple catalytic activities, and developing water-soluble or immobilized nanoreactors.
Antonina Naskalska (Jagiellonian University): Engineering of virus-like particles for modern vaccines and drug delivery vehicles
‘Virus-like particles (VLPs) are multiprotein structures that resemble viable virus particles in conformation and therefore are potent triggers of immune response in mammals. Being non-infectious and non-replicative however, VLPs are attractive candidates for safer vaccines. Interestingly, VLPs may be engineered to display heterologous proteins such as antigens or receptor targeting moieties. In addition, the cavity of some VLPs can be loaded with foreign molecules, which further extend their potential applicability. Altogether, these features render VLPs versatile delivery platforms or tools of immunization.
Here, I will present two examples of modular VLPs, where antigens of interest are anchored to the viral scaffold by two different approaches. First VLP originates from a bacteriophage and displays on its surface large trimeric antigens: Receptor Binding Domain from spike protein from SARS CoV2. Second VLP originates from a feline parvovirus, and displays linear antigens from another coronavirus (Feline Infectious Peritonitis Virus).’
3pm, 28th May 2025:
Matthew Aquilina (Dana Farber Cancer Institute, Harvard University): Crisscross origami
The crisscross polymerization paradigm has enabled the creation of the first fully-addressable hierarchical assemblies composed of thousands of unique DNA origami slats. These megastructures extend the nanoscale precision and programmability of single DNA origami structures to the micron scale, opening up new possibilities in areas ranging from cell therapeutics to molecular computing. However, their inherent complexity makes megastructure design, manufacturing and assembly an involved process. Even a basic half-micron square, which contains 64 unique origami slats, represents a major logistical challenge to build from scratch.
Since the initial unveiling of crisscross origami, the Shih Lab has been exploring various initiatives to expand the accessibility of the technique. In this webinar, I will be giving an overview of the many improvements we have made to the fabrication pipeline, which range from a solid-phase purification system, an optimization algorithm for assembly handles assignment, and a comprehensive 3D CAD package that can be used to realize new designs with minimal friction. Our ultimate goal is to refine crisscross origami into a streamlined framework for megastructure fabrication, similar to the DNA origami technique today.
Ranjan Sasmal (Arizona State University): Programming Amphiphilic DNA as GPCRs Mimic for Signal Transduction and Amplification across Membrane
G protein–coupled receptors (GPCRs) are essential membrane proteins that mediate signal transduction across lipid bilayers. Understanding the design principles of these proteins involves studying their behavior at various scales, from single-molecule biophysics to cell and developmental biology. Inspired by GPCRs function, we engineer functional membrane-spanning devices using amphiphilic DNA hairpins to transduce signals from inside synthetic and biological vesicles to produce specific responses on the outside. These DNA devices insert themselves into lipid membranes, translocating certain parts across the membrane and recognising complementary oligonucleotides on the inner side of the lipid bilayer, generating membrane fluorescence signals. Our experiments demonstrated efficient recruitment (80–90%) of Cy3-labeled targets DNA and/or RNA within cell mimetic giant unilamellar vesicles (GUVs), resulting in Cy3 halo ring formation. Upon target recognition, the DNA hairpins systematically open, partially or fully, transmitting the recognition signal across the membrane as a fluorescence output using a sequence-specific reporter dye. All-Atom MD simulation revealed the extensive stability of hairpin DNA and intermediate structures within the membrane. The kinetic features of the signal transduction process across GUVs membrane was lso evaluated by altering membrane compositions affecting the fluidity. Additionally, we employed a toehold-mediated strand displacement reaction to perform a hybridization chain reaction (HCR), amplifying the transduced signal on GUVs and live cells. Overall, our short amphiphilic hairpins may offer a general approach for non-destructive probing of cytosolic RNA biomarkers in live cells, with potential applications in transcriptomics-based isolation of target cell sub-populations and downstream therapeutic applications.
10am 24th September 2025:
Erik Poppleton (University of Munich): MD Simulations of RNA Motifs
Phospholipid bilayers are a key component of biomolecular systems, providing a barrier between the internal ‘self’ and the external environment. These molecular assemblies are formed through the self-assembly of numerous small monomers, each made up of a charged or zwitterionic, hydrophilic headgroup and a hydrophobic hydrocarbon tail. Bilayer membranes are particularly good at preventing the diffusion of large and charged molecules, such as nucleic acids, through their hydrophobic core. A 2001 paper (Vlassov, Khvorova & Yarus, PNAS 98(14): 7706–7711. (2001)), performed SELEX against DOPC liposomes and found a pair of RNA sequences, RNA 9 and RNA 10, which in complex, not only bound to DOPC membranes, but destabilized and disrupted membranes. Our interest in developing genetically-encodable cell-like molecular machinery inspires our interest in these sequences, which could allow rational design of membrane interacting and remodeling RNA origami, such as cytoskeletons (Tran et. al., Nat. Nanotech. 20:664–671. (2025)), and the goal of this work, nanopores.
While experimentally reproducing the findings of Vlassov et. al., we developed a modified version of RNA 10, which we called 10M, which was able to disrupt both DOPC and DPhPC membranes without RNA 9, simplifying the positioning and stoichiometry problems which using two interacting aptamers presented. However, despite this success, it is still unclear how this class of membrane-affine aptamers function. Hence, we turned to molecular dynamics (MD) simulations to investigate the molecular basis of recognition of membrane surfaces by these sequences.
Here we present the results of these simulations, showing that RNA sequences are able to form hydrogen bonding networks with choline headgroups, resulting in contacts between nucleobases and the membrane core and thinning of the lipid bilayer. Based on these results, we seek to rationally design this novel RNA behavior, which will both add a new tool to molecular nanotech, and may elucidate recently-discovered membrane-associated RNA sequences (Lyon & Breaker, RNA, in press (2025)).
10am 29th October 2025:
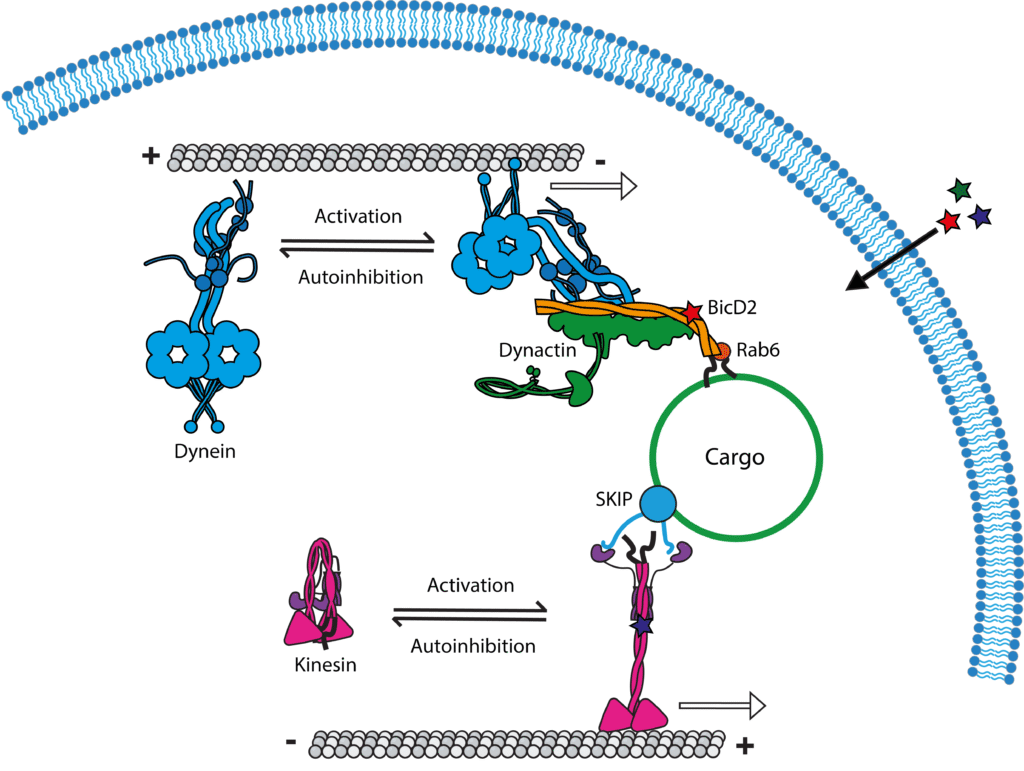
Jessica Cross (University of Bristol): Hijacking Hinges: Cracking the coiled-coil code to control intracellular transport
Cytoskeletal motor proteins are nature’s nanoscale machines, driving the organisation and transport of cellular cargo. Their activity is tightly regulated by dynamic structural switches that toggle between inactive and active states. My research integrates rational protein design, synthetic biology, and AI-based modelling to uncover and engineer these molecular “hinges,” focusing on coiled-coil domains that act as conformational switches. Recently, we discovered and reprogrammed a coiled-coil hinge in the motor kinesin-1 to create an inducible ON/OFF switch for motor activity in living cells. These synthetic peptide tools provide direct control over intracellular transport and open routes to engineer programmable molecular machines. Ultimately, this work aims to define the principles of conformational regulation and harness them to build responsive protein systems for applications in synthetic biology and nanomedicine.
10am 26th November 2025:
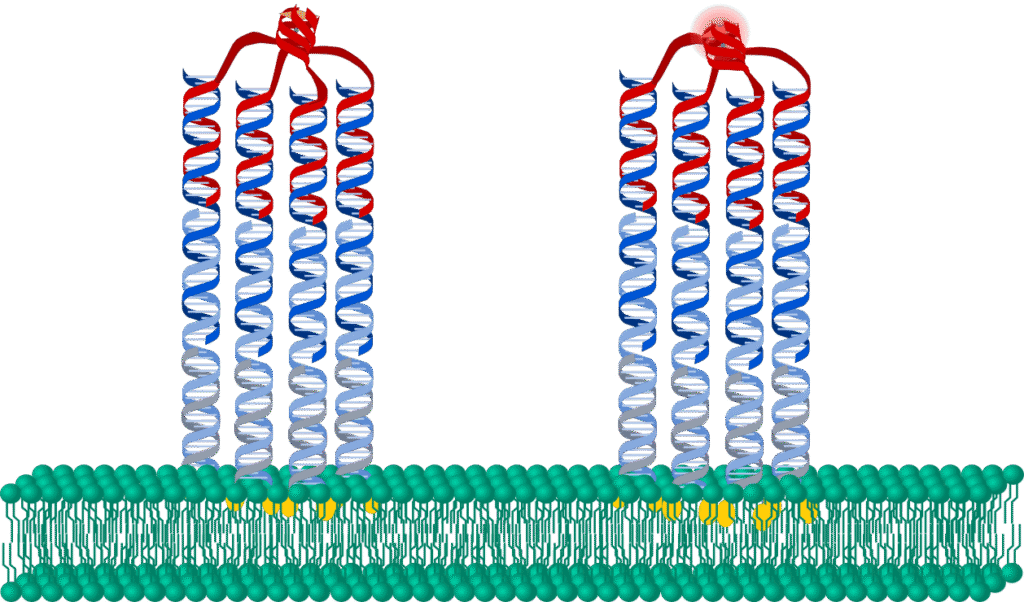
Roger Rubio Sanchez (University of Cambridge): Biomimetic DNA-based receptors for bottom-up membrane functionality in synthetic cells
Biological membranes regulate the spatio-temporal organisation of their protein machinery to facilitate key functionalities, like signal transduction, molecular trafficking and cellular motion. Bottom-up synthetic biology aims to construct life-like behaviours in synthetic cells, allowing us to study biological phenomena and to develop potential applications in biomedicine, biosynthesis, and bioremediation [1].
My work applies the tools of bionanotechnology, from self-assembly to chemical nanotechnology, to engineer functionality in otherwise inert lipid bilayer membranes [2-5]. Here, I will discuss the design and application of DNA nano-devices with controlled structure, catalytic activity, and membrane distribution [5]. Besides fundamental contributions to our understanding of biological membrane organisation and activity, our biomimetic receptors pave the way for sophisticated sensing, communication, motion, and division pathways in synthetic cellular agents.
References:
[1] Acc. Chem. Res. 50, 769–777 (2017).
[2] Chem. Commun. 57, 12725 (2021).
[3] Nano Lett. 21, 2800–2808 (2021).
[4] J. Am. Chem. Soc. 145, 11265–11275 (2023).
[5] J. Am. Chem. Soc. 147, 33780–33789 (2025).
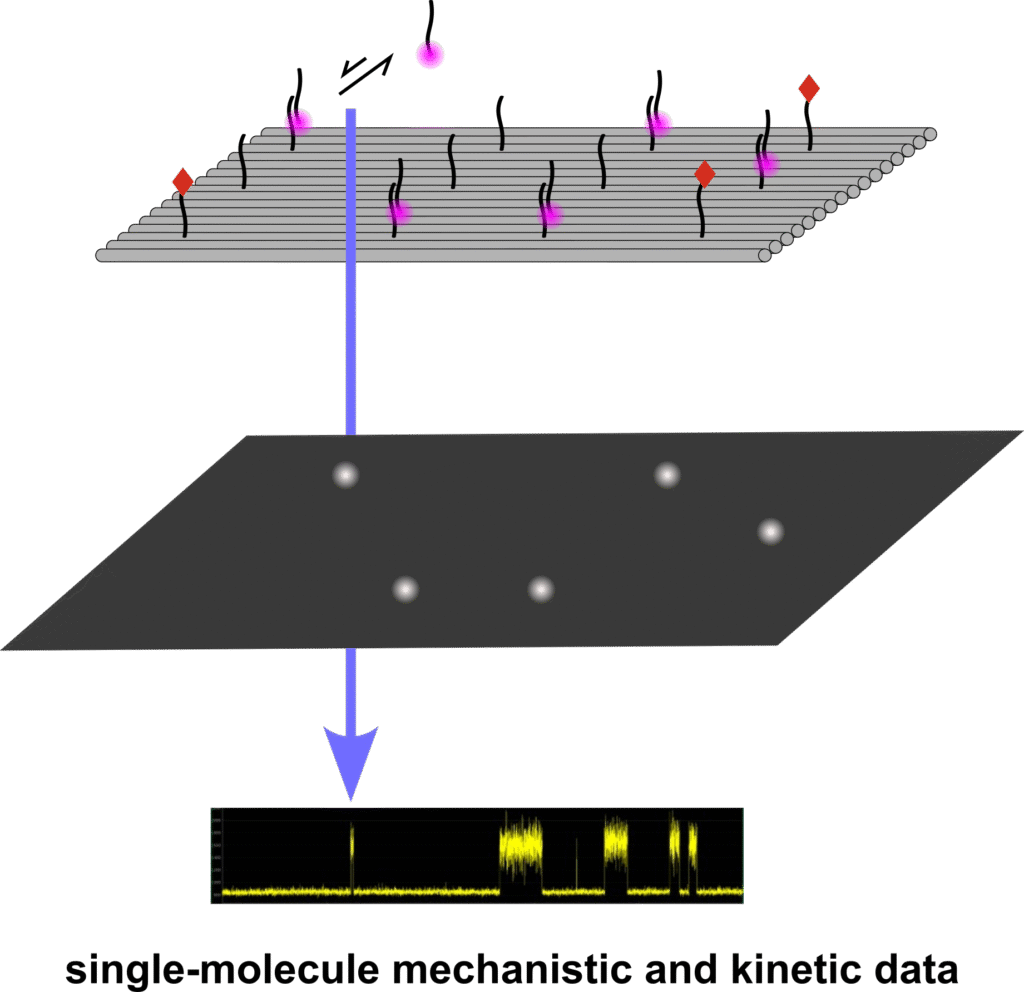
Luca Piantanida (University of British Columbia Okanagan): Molecular nanoscale detection on DNA pegboards for single-molecule kinetic and interaction analysis
Engineering DNA nanostructures by optimizing the fine equilibrium between shape, architecture, and functional activity enables the development of intelligent bio-designs for molecular nanometrology. Here, DNA nanotechnology is used to construct DNA pegboard platforms that organize molecules with exceptional nanoscale precision, organization, and spatial control. These platforms provide a highly controlled environment for molecular nanoscale detection, facilitating single-molecule mechanistic and kinetic analyses. By enabling such investigations at the nanoscale, this technology drives a detailed understanding of molecular interactions of small molecules and their potential as smart therapeutics.
10am 28th January 2026:
Nils Winkler (Heidelberg University): Genome condensation in Sars-CoV-2: using math to understand viral assembly
While many enveloped RNA-viruses use a rigid protein capsid to package and protect their genome, SARS-CoV-2 uses a high density of the transmembrane protein M to structurally reinforce its membrane, which then bends around the viral genome condensed by the cytosolic protein N. Interestingly, the RNA genome seems to be further subdivided into smaller subunits, leading to a nested egg structure. We theoretically study the energetics of this situation by minimising appropriate energy functions for genome condensation, membrane bending, droplet wetting and wrapping. In particular, we investigate which of these energy contributions can explain the nested egg structure as observed experimentally.
10am 25th February 2026:
Oliver Dennis (EPFL Lausanne): Evolving a human protein cage for RNA packaging
A major challenge in mRNA delivery is getting nanocarriers to efficiently package RNA cargo. In this project, we use protein engineering and directed evolution to repurpose an endogenous human protein cage for improved mRNA packaging. We first engineered protein cage variants that can package their own encoding mRNA. We then generated mutagenised libraries which are are expressed and purified, and functional selection is applied by enriching for RNase-resistant assemblies, allowing us to select for improved RNA packaging and protection. Further rounds of evolution will be guided by long-read sequencing and cryo-EM structural analysis, which will help us connect sequence changes to packaging efficiency and structural effects. This work aims to establish a basis for developing non-viral protein-based RNA delivery systems.
Us and our supporters





