A year on, what’s changed – Kirstie Morrice [Reposted from ECRF Genetics Core Blog]
2021 here we are. How has your working life changed over the last year?
Here at the Genetics Core, like so many other workplaces across the UK, we closed our doors in March 2020. A surge of planning began; how do we work from home? What is expected of us? How do we adapt our laboratory jobs? Although contingency planning had begun well before we went into lockdown, I don’t think it really prepared us for what lay ahead. Our meetings had gone from all of us onsite to people dotted around living rooms, kitchens, hallways and staircases. The dreaded mute/unmute fiascos had started, children curious to see what mum or dad were doing, pets greeting the postie, the list goes on and on.
As we settled down into our work from home lives we set our sights on reviewing, updated and writing our Standard Operating Procedures (SOPs), Risk Assessments (RAs) and Control of Substances Hazardous to Health (COSHH) assessments – a task that was always pushed aside as we would run off into the lab with purpose!
Using our time to focus on the Core development; researching automation for both our sample processing team and sequencing team, website development, continuing the development of the Genetics Core’s improved training competencies. The ‘Tech Session’ was created and technicians were asked to present protocols to the team in order to reaffirm their learning while educating others. This time allowed technicians to work together to make informative and interactive learning experiences for the whole team, creating a document that would allow trainees to look to for a clear understanding of a process.
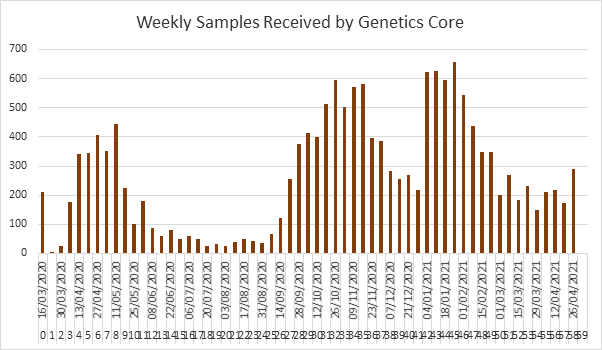
As we all worked away, the front line staff were hard at work helping to save lives. Our Research Nurses moved to front line patient care, while Professor Kenny Bailie, an intensive care Doctor, expanded his already existing GenOMICC project (Genetics of Mortality in Critical Care). The GenOMICC project was created to allow research of future pandemics and had all of the ethics and SOPs in place to rapidly allow a greater understanding of critical illness in COVID patients. At this point, in March 2020, we had 100 samples from patients. As the project opened up to COVID sufferers in critical care, the numbers of research samples grew rapidly, with Intensive Care Units (ICU) across the UK signing up to help contribute to the ongoing medical research effort. For more information about the GenOMICC project, please visit their website here; GenOMICC.
This project, that was sitting as a 100 sample project, was to turn into the biggest project the Genetics Core has ever work on, with an estimate of 10,000 samples in the beginning. The uncertainty of when we would be back in the lab was now dependent on the approval of the Risk Assessment allowing us to start processing the Whole Blood of COVID positive patients for genetic analysis.
As the risk assessment was approved, three weeks after lockdown the doors to the Genetics Core reopened and we had very little time to think, we had to act. We adapted quickly and efficiently and began processing the highest number of samples possible per day, per week. This is where the Genetics Core began its new journey, the personal and professional growth of each technician over the next 12 months contributed to the growth of the Core as a whole. As we reflect on how much has changed it truly is a testament to how hard every single person worked to create this new version of the Genetics Core.
The Sample Processing Journey; Are we being replace by robots?
The sample processing team at the Genetics Core is made up of 5 technicians; Sarah McCafferty (RSci), Alan Maclean, Kataryzna Hafezi (RSciTech), Kirstie Morrice (RSciTech) and our latest edition to the team Lorna Donnelly.
As a team we offer a variety of services ranging from nucleic acid extractions to peripheral blood mononuclear cell (PBMC) isolation. This past year has been unlike any other, with all clinical trials paused, samples from other projects halted and the 5 of us focussed on one project, one protocol at the one time, our team banded together to take on the Genetics Core’s biggest project to date.
Deoxyribonucleic acid (DNA) extraction is the process by which DNA is separated from proteins, membranes, and other cellular material contained in a cell. Our manual protocol uses the Nucleon BACC3 commercial kit to extract good quality DNA and is made up of four steps including; cell lysis, DNA deproteinisation, DNA precipitation and DNA cleaning. Extracted DNA is used in downstream processing across the Genetics Core facility. This protocol has been used at the Genetics Core for over a decade, it’s a tried and tested method that allows us to extract high quality and quantity DNA for research groups all over the world.
As we look back at how we started the year off, we were so focussed on high productivity, we didn’t have the luxury of stepping back to assess the situation, we were learning how to process and manage these numbers of samples as the weeks went on. The options to adapt and move towards automation had been discussed, but were met with hesitation at first.
Is it really worth it?
I can do it in the same time!
I don’t trust it…
So, what is it about automation that makes it worth it?
In order to understand the changes it has made for our team, we need to dive into our DNA Workflow.
As with any manual protocol, there is a lot of hands on time required. This was something that saw the sample processing team stretched to capacity when required to do extractions while; confirming safe receipt of hundreds of samples per day, maintaining stock levels of consumables and reagents for the extraction process to continue. Using the 4, 5, 30 minute breaks in protocol to do housekeeping tasks, with reduced numbers on site, every minute counted! Sounds exhausting right?
So what happened when our tried and tested manual protocol was put to the test with the overwhelming number of samples. Well, as predicted, good quality and quantity of DNA was extracted. Yes! What did you think would happen? Although the kit was performing as it has been for decades, the variables that saw us looking to adapt were the possibility of less hand on time and a quicker turn around processing time.
To put this into perspective, as per our manual extraction protocol, our newly extracted DNA aliquots were set on rotary wheels for two weeks to resuspend in solution. As our sample numbers grew and grew, the process of waiting for two weeks before quality checks became too long. The samples kept arriving, in recording breaking numbers, we were running out of rotary wheels and we needed to get samples sent for sequencing at Genomics England, who are partnering with GenOMICC providing whole genome sequencing (WGS).
Our workflow overview;
DNA Aliquot – 2-week rotary wheel – split aliquot; 500uL in Deep Well Plate & 500uL in 1.5ml microtube – QC DW Plate – Normalisation
Platforms for automating our nucleic acid extractions were discussed, the Perkin Elmer Chemagic 360 became the front runner for us, with the ability to extract up to 10mL blood. The Chemagic 360 offers different rod head compatibility, 12, 24 and 96 sample rod head, determined by sample volume input, sample type and sample throughput.
We opted for the 12 sample rod head as it fitted in with our current protocol. Although the time of the run was not dramatically faster than manual extractions, it did allow the technician running the platform to be free to help with sample receipt, housekeeping and set up consumables for the next run. The 12 sample rod set up still allowed us to continue with our DNA workflow but with one new revelation, the DNA is resuspended in elution buffer and QC can be performed immediately after processing. By implementing this, the 2-week rotary wheel step had been cut from our workflow;
DNA Workflow update;
DNA Aliquot – Split Aliquot, 500uL in Deep Well Plate – QC DW Plate – Normalisation
For us at the time, it was an incredible change that made the workflow more streamlined than ever before. As we continued to plough our way through the samples we were now on the second big wave of cases that hit across the UK. As ICUs became overwhelmed with cases, the numbers continuing to grow and the marvels of funding, we were bought two additional Chemagic 360 platforms. This saw us upgrade from one 12-rod head to having three 24-rod head platforms, allowing 72 samples to be processed at one time across the three platforms. In January 2021 we added two more Chemagic 360 platforms to the team, welcome Bert and Ernie! Now all three platforms were fitted with the 24-rod head, which saw our sample throughput increase exponentially.

K.Morrice, L.Donnelly and A.Maclean pictured with Chemagic 360 Platforms.
We were now processing 192 samples per day, allowing us to cut down our backlog of samples and have the capacity to process the daily arrivals! Looking back now, it is quite unbelievable the difference in our workflows, how we work as a team and as always, the numbers do not lie. As we adapted and figured out the best way of working with our new platforms, we settled into our new way of working as if we’d been doing it for years, even though we were only approaching a year of COVID research service and a month of our Chemagic 360 24-rod head workflow. The month after installation, February 2021, we processed more samples than we did in the whole of 2019. There are some things you just cant go back to and I think I speak for all of us when I say, the automation of the DNA extraction workflow has been an eye-opening experience, but ultimately we wouldn’t go back!
As explained when we saw our 12-rod head Chemagic 360 installed, the upgrade to the 24-rod head brought with it further streamlining of our workflow.
DNA Workflow update;
Eluted DNA straight to Deep Well Plate – QC DW Plate – Normalisation
The 24 rod head sample volume input was a maximum of 4mL, meaning the eluted DNA was a maximum of 400uL. Previously, we were eluting 1000uL and storing 500uL in a deep well plate and 500uL in a microtube. Immediately, we saw a reduction in consumables as the 24-rod head Chemagic 360 allowed us to elute the DNA straight into a deep well plate. Not only did this save us hands on time splitting the aliquots, it has saved storage space in our freezers!
Now, this may all seem like a rave review for the Chemagic 360 platforms, and yes, I guess it is. There are parts of the platform that are worth thinking about; sustainability, the Chemagic 360 platform seemed very plastic heavy, but let’s compare manual and our new automated counterpart;
Manual:
- 15mL/50mL Falcon tubes
- 1.5mL microtubes
- Pastettes
- Pipette tips
- Deep Well Plate
- Energy used by running the autoclave to sterilise consumables
- Energy used by storing DNA in Freezers; tubes and plates
- Energy used by storing EDTA tubes in Freezers, collecting backlog samples from freezers
Automated:
- Plastic consumables Kit
- 12mL tubes (elution/waste tips)
- Magnet Tips
- 24 well plates
- Pipette tips
- Deep Well Plate
- Energy used to store DNA; halved
- Energy used to run platforms
As we strive to meet better sustainability standards, here at the Genetics Core we hold a Gold award for our efforts in sustainability but we are always looking to see what areas we can improve on. Although the level of plastic consumables used may not change, I can’t help but highlight the reduction in storage that is required now we run the 24-rod head. This has already been rolled out across other projects within the Genetics Core.
So, ‘is automation worth it?’, it was worth it for us.
This last year, we have upgraded our working practices, streamlining our workflows, reducing hands on time, creating a more sustainable working environment for our team. It will allow us to work on projects that may once have seemed huge, 10,000 samples manually? At our current capacity, that level of work is achievable in months!
The success the sample processing team has had with integrating an automated platform into our workflows has now paved the way for the Genetics Core to examine what other workflows can be streamlined – what’s next for our team of 12?
Before we jump ahead to the future, lets have one last moment of reflection. I approached the team and asked them, quite simply;
‘What 3 words would you use to describe our last working year?
Around this time every year, we see annual appraisals taking place across the University. Taking advantage of the already reflective mood, I asked everyone at the Genetics Core to describe the last year in three words, I think the results speak for themselves.

Our team has endured and thrived through hard work and determination, but most importantly we worked as a team. As a result we see a new streamlined version of the Genetics Core, what will the next year bring? Stay tuned to see how our wee team continues to grow.
Kirstie Morrice is a sample processing technician working at the Genetics Core. Kirstie enjoys staying active with yoga, bouldering and walking Munros around Scotland. Kirstie can be contacted at kirstie.morrice@ed.ac.uk.




Recent comments