Cell vs Virus – the (in)finite battle between two enemies
As a PhD student from the Institute of Genetics and Molecular Medicine in Edinburgh, I had the great opportunity to attend the talk “The epigenetic transcriptional repressor complex – defending the genome from retroelement attack” given by Prof. Paul J Lehner, a Wellcome Trust Principal Fellow from the Cambridge Institute for Medical Research.
Prof. Lehner and his lab group focus on the particular question: How are viruses recognised by the cellular machinery and subsequently eliminated? And in turn: how do viruses overcome these defensive mechanisms that result in viral infections of the cell? Investigating this question would not only help us to understand how viruses manipulate the host cell, but would also provide new strategies and approaches in the therapeutic treatment of viral diseases– such as HIV or Hepatitis B.
At the beginning of his talk, Prof. Lehner gave insights into the characteristics of viruses and illustrated a cascade of steps a virus will undergo that lead to infection. Since viruses lack replication machinery of their own, they are forced to hijack machinery belonging to other cells in order to reproduce and thrive. Firstly, genomic material in the form of RNA or DNA is introduced into the host cell which allows the virus to recruit and utilise the machinery of its host. This leads to viral genome replication and spreading throughout the host body causing the viral infection. To defend themselves against viral attack, organisms have adapted their immune system to recognise and destroy foreign DNA. Nevertheless, viruses developed different strategies to overcome these immune responses. For instance, many viruses such as Hepatitis B, remain in a silent state until they are stimulated to reactivate. On the contrary, retroviruses are unique and integrate their DNA into the host genome and essentially become “genes”. Consequently, the treatment of viral illnesses such as HIV is very challenging with there being no known cure, thus current clinical treatments must be accompanied by lifelong therapy.
One critical step towards efficient treatment is to understand the signalling pathways during viral infection by revealing novel genes that could be targeted.
Prof. Lehner presented one of these novel targets – the HUSH (human silencing hub) complex – which comprises of the three proteins TASOR, MPP8 and Periphilin – was identified by a haploid genetic screen1. The HUSH complex is bound at H3K9me3-rich regions of the genome and plays a crucial role in heterochromatin maintenance. HUSH performs this role by the recruitment of the histone lysine methyltransferase SETB1 which results in the heterochromatic spreading across its target sites mediating the epigenetic repression of newly integrated transgenes1. Since HUSH is found primarily at methylated sites at the genome, the Lehner lab wanted to investigate in more detail how chromatin state impacts HUSH function. Through a genome-wide CRISPR-screen, the gene MORC2 was identified as an epigenetic regulator required for chromatin remodelling and condensation, which is a prerequisite for HUSH-mediated gene silencing2. By making use of the DIVA technique, recently developed by the Lehner lab, they further explored the relationship between MORC2 and chromatin compaction. DIVA (differential viral accessibility) is a powerful method that allows the genome-wide discrimination between “open” (accessible) and compacted (less accessible) chromatin by using lentiviral vectors that are transduced into cells followed by integration site analysis3. Prof. Lehner elucidated the advantages of DIVA compared to the currently used methods such as ATAC-Seq. that are strongly biased towards the detection of certain regulatory regions. DIVA overcomes this limitation and enables the profiling of a broader range of integration sites including those at gene bodies and intergenic regions. Prof. Lehner demonstrated the DIVA method by comparing chromatin structure of wildtype and MORC2knockout cells and showed that loss of MORC2 resulted in increased viral accessibility due to chromatin decompaction. This occurred concomitantly with the loss of HUSH activity that led to transcriptional derepression2. This underpins the importance of HUSH-mediated silencing with regards to latent retroviral genes.
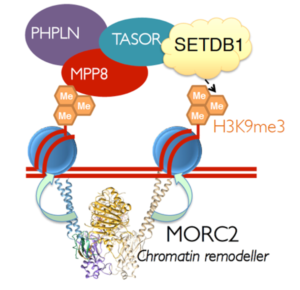
Structural model of essential factors that are required for transgene silencing. The HUSH complex is formed by Periphilin (PHPLN), TASOR and MPP8 and binds to the repressive histone modification mark H3K9me3. The HUSH complex recruits MORC2, a chromatin remodeller, and SETB1, a histone methyltransferase, to heterochromatic sites which mediates transgene silencing. © 2019 Prof. Paul J Lehner All Rights Reserved
Viral latency – a state where viral transgenes lie dormant in the host genome until reactivated by a stimulus – in heterochromatic regions is a huge issue, being responsible for the recurrence of infection in many cases. The crucial discovery of the link between the HUSH complex and the regulation and maintenance of heterochromatin could prove to be a massive step forward in the fight against viral latency.
I was very fascinated by Prof. Lehner’s talk and the great work they have done that led to the identification of novel factors involved in viral pathogenesis. Further studies of the identified genes could shed light on the pathways that viruses use to infect their host cells. This provides great clinical potential in enhancing treatment of several viral diseases. Furthermore, the talk was additionally beneficial since Prof. Lehner showed a broad range of applications for the DIVA method, that paves the way for enhanced structural chromatin studies.
1Timms, R. T., Tchasovnikarova, I. A. & Lehner, P. J. Position-effect variegation revisited: HUSHing up heterochromatin in human cells. BioEssays38, 333–343 (2016).
2Tchasovnikarova, I. A. et al.Hyperactivation of HUSH complex function by Charcot–Marie–Tooth disease mutation in MORC2. Nat. Genet.49, 1035–1044 (2017).
3Timms, R. T., Tchasovnikarova, I. A. & Lehner, P. J. Differential viral accessibility (DIVA) identifies alterations in chromatin architecture through large-scale mapping of lentiviral integration sites. Nat. Protoc.14, 153–170 (2019).
(© 2019 Prof. Paul J Lehner All Rights Reserved)



